Digital PCR, bloom right now! dPCR development history In 1992 , Sykes et al. identified mutant leukemia cells from non-lymphocyte and normal somatic cell backgrounds and tested low-abundance IgH heavy chain mutant genes in complex backgrounds. Three important principles were proposed in this study: limited dilution, presence or absence of endpoint signals, and statistical processing of the Poisson distribution of data, which laid the foundation for the development of dPCR. In 1997 , the study used a glass capillary as a carrier for PCR reaction, which can quantify the template molecule, and verified that the Taq-Man probe can detect template single molecules, especially in a small reaction system, and developed a single molecule quantitative technique. dPCR single template amplification provides technical support. In 1999 , Bert Vogelstein and Kenneth W. Kinzler formally proposed the concept of dPCR. When detecting KRAS mutations in the stool of colon cancer patients, they first assigned the samples to 384-well plates, and then used different fluorescence to detect the mutant genes and normal. The gene, the mutation rate is obtained by calculating the ratio of the mutant gene to the normal gene. In 2003 , Dressman et al. invented a BEAMing method (beads, emulsions, amplification and magnetic properties). BEAMing separates the mixture of template, primer, PCR reagent and magnetic beads into droplets, most of which contain no or only one template. After the PCR is completed, the amplified product will be linked to the magnetic beads by the binding of biotin-streptavidin, and the emulsion will be broken up, and the beads will pass through the detection oligonucleotide and flow. The hybrid of the cytometer is read. In 2006 , Fluidigm introduced the first chip-based commercial digital PCR system. Subsequently, QuantaLife developed microdrop digital PCR technology using water-in-oil droplet generation technology. In 2011, QuantaLife was acquired by Bio-Rad, and its microdrop digital PCR instrument was renamed as QX100. In 2013, the company introduced the upgraded model QX200. In the second half of 2013, Life-technologies introduced QuantStudio 3D digital. PCR system. Overview of commercial digital PCR platforms Fundamental The principle of dPCR is to distribute the standard PCR reaction system containing nucleic acid template evenly into tens of thousands and millions of PCR reactions, and distribute them into chips or droplets so that each reaction contains as many template molecules as possible. Molecular template PCR reaction, using a labeled probe to detect a specific target sequence, by reading the presence or absence of a fluorescent signal, calculating the ratio and number of reaction units of the two signal types, and performing absolute quantification by calibration of the statistical Poisson distribution. The initial copy number (concentration) of the sample can be obtained based on the total number of digital PCR reaction units and the number of units with fluorescent signals and the dilution factor of the sample. Unlike qPCR, which performs real-time fluorescence measurement on each cycle, the dPCR technique collects the fluorescence signal of each reaction unit after the end of amplification, and finally derives from the Poisson distribution principle and the number and proportion of positive droplets. The starting copy number or concentration of the target molecule. According to the implementation of a large number of independent reaction units, dPCR is subdivided into droplet microfluidics based droplet digital PCR (ddPCR) and microarray microchip based on chip microfluidics. (chip digital PCR, cdPCR). Microfluidic chip digital PCR With the development of microfluidic chip technology, digital PCR can accurately and quickly divide a sample into several independent reaction units by means of a microfluidic chip. Each unit does not affect each other, and multiple steps can be performed simultaneously. Increasing the reaction throughput also reduces the cost of reaction consumption. Currently Fluidigm's Bio-Mark TM Genetic Analysis System, Life Technologies company QuantStudio TM 3D digital PCR PCR systems are digital systems are microfluidic chip technology. Droplet digital PCR Digital PCR is derived from emulsion PCR technology. It can generate tens of thousands or even millions of nanoliters or even individual water-in-oil droplets at a time as a sample dispersion carrier for digital PCR. The fluorescence signal of each droplet was detected after the reaction was completed. At present, Bio-Rad's QX100 system and QX200 system are digital PCR systems using droplet technology. Comparison between digital PCR and real-time fluorescent quantitative PCR application dPCR technology has the following advantages: (1) High sensitivity. dPCR essentially transforms a traditional PCR reaction into tens of thousands of PCR reactions, and independently detects the target sequence in these tens of thousands of reaction units, thereby greatly improving the sensitivity of the detection. (2) High accuracy. By calculating the number and proportion of positive reaction units in tens of thousands of reaction units, dPCR can accurately detect differences in the sequence of interest with little change. (3) High tolerance. The first step of the dPCR technology is to distribute the background sequence and the PCR reaction inhibitor to each reaction unit, and most of the reaction units do not contain the target sequence, and the low-abundance target sequence is relatively rich. It is concentrated in some reaction units, thereby significantly reducing the interference of background sequences and inhibitors in the reaction units. In addition, dPCR only judges the positive/negative state when the result is judged for each reaction unit, and does not depend on the Ct value, and is greatly affected by the amplification efficiency, and the tolerance to the background sequence and the inhibitor is also greatly increased. improve. (4) Absolute quantification. PCR directly calculates the copy number of the target sequence, allowing accurate absolute quantitative detection without relying on Ct values ​​and standard curves. The above advantages of dPCR make it particularly suitable for applications in the following areas: As a new technology for nucleic acid quantification, dPCR has higher sensitivity and accuracy than previous nucleic acid quantification methods. dPCR provides new detection methods and experimental ideas for molecular biology, microbiology and other fields. From the perspective of application range and experimental cost, dPCR cannot completely replace qPCR, but dPCR method can perform accurate absolute quantitative analysis, excellent data reproducibility, and low sample demand, in nucleic acid detection and quantification. There are very important additions to the aspect. In the development process of the whole dPCR, the application is more and more extensive, and it has important significance in the quantitative detection of nucleic acid, monitoring of antiviral therapy and prognosis. With the development of dPCR technology and commercial platform, dPCR will have higher throughput and lower cost, and will play a more important role in scientific research. references Medical Equipment Disposal,Syringes Needles Sizes,Disposable Syringe,Insulin Syringe FOSHAN PHARMA CO., LTD. , https://www.foshanpharma.com Types of supplier Production time Instrument model Number of liquids Characteristics Chip type Fluidigm 2006 EP1 36,960 Based on an integrated fluid path chip, the fluid is quickly and accurately divided into individual units. The operation is time consuming and the reaction cost is high. 2006 BioMark HD 9180 Life Technologi-es 2009 Open Array 3072 The number of reaction repeats can be easily adjusted to meet application needs.
The samples are completely isolated to prevent cross-contamination. The flux of the reaction is low. 2009 QuantStudio 12K Flex 3072 2013 QuantStudio
3D 20,000 Microdrop type Bio-Rad 2011 QC100 20,000 Based on water-in-oil technology, a large number of droplets provide sufficient accuracy to accurately determine sample copy number. 96 sample reactions can be performed simultaneously. 2013 QX200 20,000 RainDance 2012 RainDrop 10,000,000 The skin upgrade reaction droplets provide a higher detection dynamic range and are suitable for processing different samples with larger concentration differences. 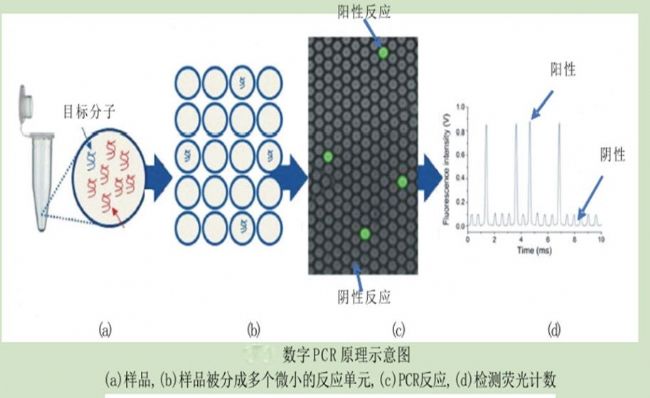
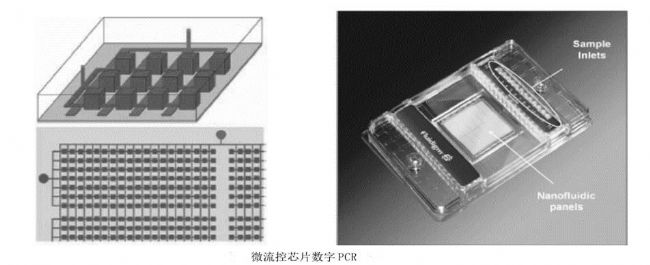
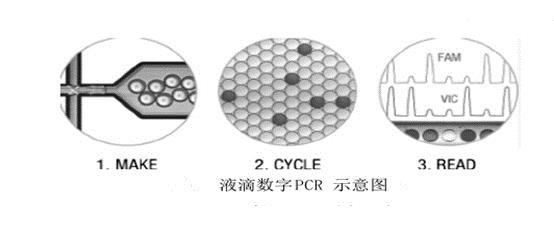
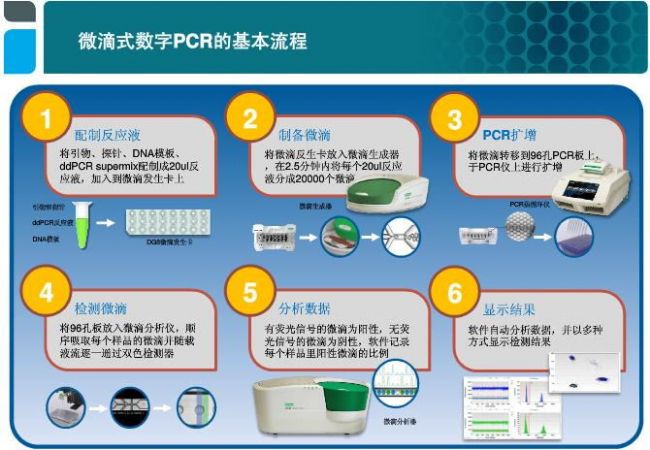
method principle application advantage Disadvantage Digital PCR The PCR reaction system is divided into liquid and PCR amplified. The number of copies of the sample is directly obtained by counting the number of positive and negative reactions. Absolute quantification of viral load; copy number variation analysis; detection of rare mutations; quantification of gene expression; analysis of second-generation sequencing libraries, etc. Absolute quantification, no standard curve required, high sensitivity, better tolerance to PCR reaction inhibitors The dynamic range of detection is small, it is easy to produce false positives, and the cost is high. Real-time PCR The amount of amplified product is proportional to the intensity of the fluorescent signal, and the sample is quantified by the cycle threshold of the reaction and the standard curve. Detection and quantification of pathogens; relative detection of gene expression; single nucleotide polymorphism analysis; detection of tumor markers, etc. Wide dynamic range of detection, wide application range and low cost Amplification efficiency is susceptible to PCR inhibitors field Applications clinical diagnosis Non-invasive prenatal diagnosis Cancer marker detection Virus detection Copy number variation analysis Rare mutation detection Gene expression analysis Complex sample gene expression analysis Single cell gene expression analysis Next generation sequencing Sequencing results verification Sequencing library quality control Quantification of genetic components Genetically modified component analysis Microbial Detection Water sample microbial detection Pathogenic microorganism detection